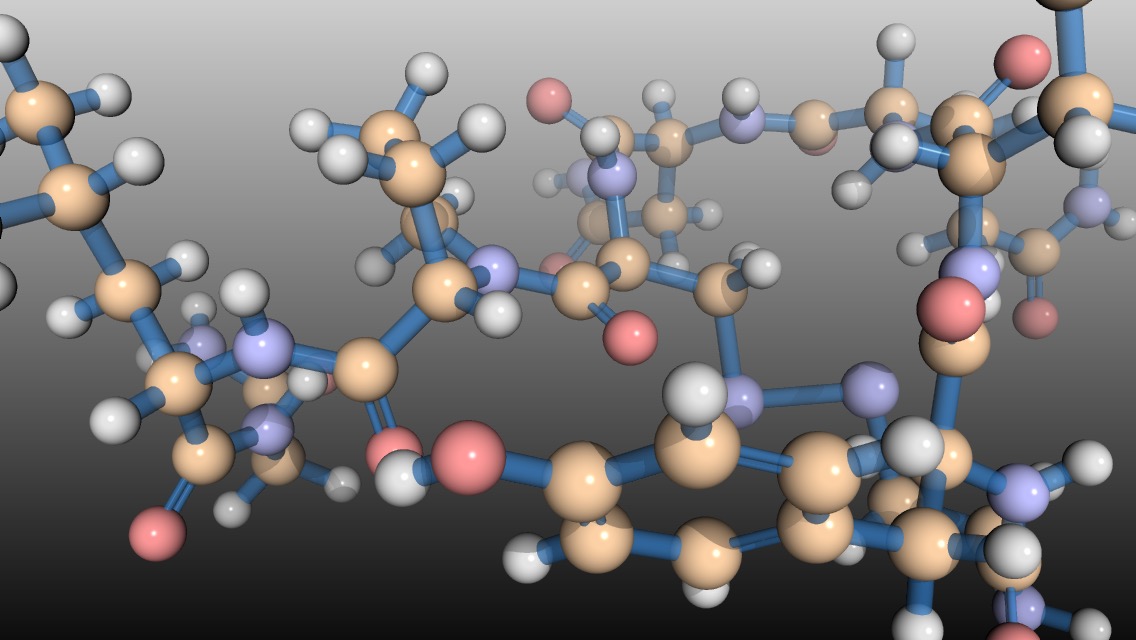
I often show molecular structures when I teach the biochemistry section of Integrated Principles of Biology, and the typical 2D representations often obscure some of the key 3D features. With the help of Pymol, ChemSpider, and FFmpeg, I can easily make video of molecular structures where the molecule rotates. This helps demonstrate the inherent 3D properties of the molecules and looks good, too. Here is a quick tutorial.
- First, choose an interesting small molecule. For this example, I chose Oxytocin, also known as the “love hormone”.
- Open your browser and go to ChemSpider. Search for Oxytocin.
- Click the
3Dbutton under the image of the structure of Oxytocin. - Click the
save to diskbutton to save the 3D structure file to your disk. I renamed the388434.molfile toOxytocin-388434.mol. - Start PyMol. Open the 3D structure file (with the
.molextension) in PyMol.
At this point, you can manipulate the structure file as you would any other structure in PyMol. Below I have included a PyMol script that will apply nice styling to the molecule, but you can style the molecule however you like. This script also contains the movie commands, so comment out the movie lines if you just want to play with the molecule styling or you will have to wait while PyMol tries to ray trace every frame.
PyMol has several utilities to make simple movies, and the script below uses the util.mroll utility. If you have a PyMol subscription, you can save the movie directly as an mpeg, but I compiled my own version of PyMol so that function is missing. Nonetheless Pymol will save each frame of the movie as a .png file and I can create a movie from the files using the free program, FFmpeg. There is a learning curve with FFmpeg, but there are lots of resources available. For some FFmpeg encoding tips, see the post by Jess Hyden on Octoshape Support and the FFmpeg wiki. In the PyMol script, below, I have included the FFmpeg settings that I used to generate the movie in this post.