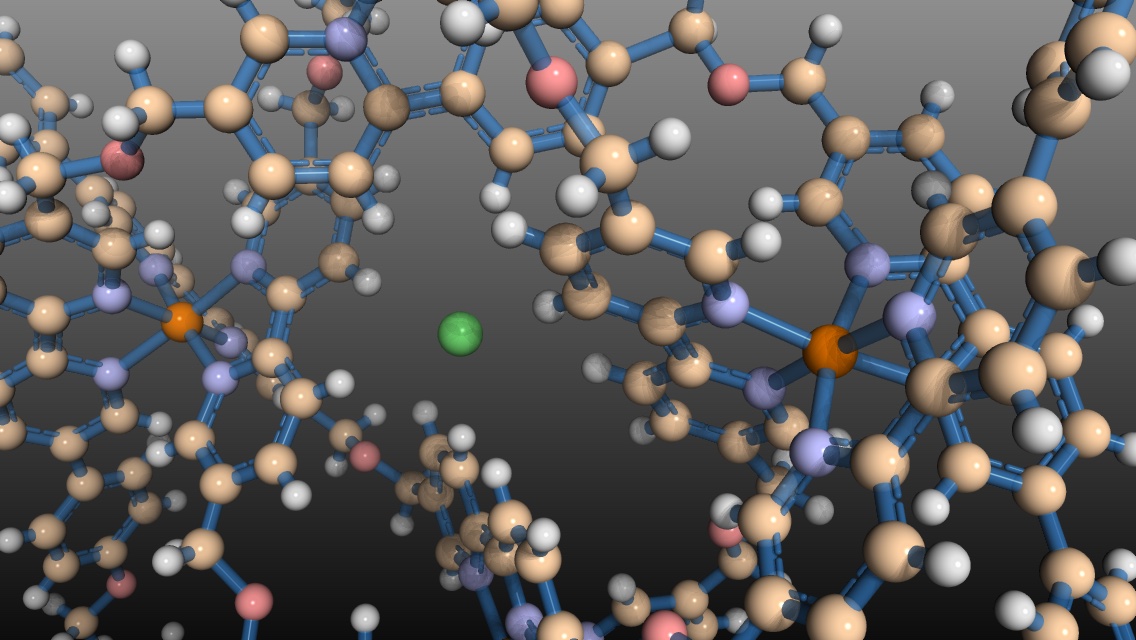
molecular knot from David Oppenheimer on Vimeo.
I read an interesting article about the design and synthesis of a molecular knot in Science by Danon et al.1, and I thought I would try to play around with the molecule in the molecular structure manipulation program PyMol. Here’s how I made the movie shown at the beginning of this post.
-
Get the structure file. The paper mentioned that they deposited the X-ray crystallography structure data in the public Cambridge Structural Database (CSD) under the Refcode entry, ODEQIA (CCDC Number, 1497422). I downloaded the structure file, which was in
.cifformat. -
Convert the file to
.pdbformat. The Mercury software package can convert.ciffiles to.pdbformat. Download Mercury, install it and open the.ciffile. Go toFile -> Save As...and choosePDB filesforFiles of type:. Give your file a name and clickSave. -
Open the
.pdbfile in PyMOL. Once you open the file in PyMOL, you have myriad possibilities for viewing and animating the molecule. -
Format the molecule. I like to use a ball and stick format for small molecules so that I can see all the atoms. I have previously found a ball and stick color scheme that I like on the PyMOLwiki (one of the random images in the upper left). To format the molecule and make the movie shown at the beginning of this post, you can use the PyMOL script below. See the comments in the script for details.
-
Write the movie files. Either uncomment the
mpngcommand before you run the scrip, or typempng name-of-file-prefixto write your movie frames to a series of.pngfiles. -
Make the movie using
ffmpeg. If you purchase a license for incentive PyMOL, you can save a movie from within PyMOL proper. Since I am using open-source PyMOL, I use the free program, ffmpeg to make my movies from the series of.pngfiles produced by PyMOL. The settings I used are included in the script below.
Molecular Knot Movie
-
Danon, J.J., A. Krüger, D.A. Leigh, J.F. Lemonnier, A.J. Stephens, I.J. Vitorica-Yrezabal and S.L. Woltering. 2017. Braiding a molecular knot with eight crossings. Science 355: 159-162. link to article ↩