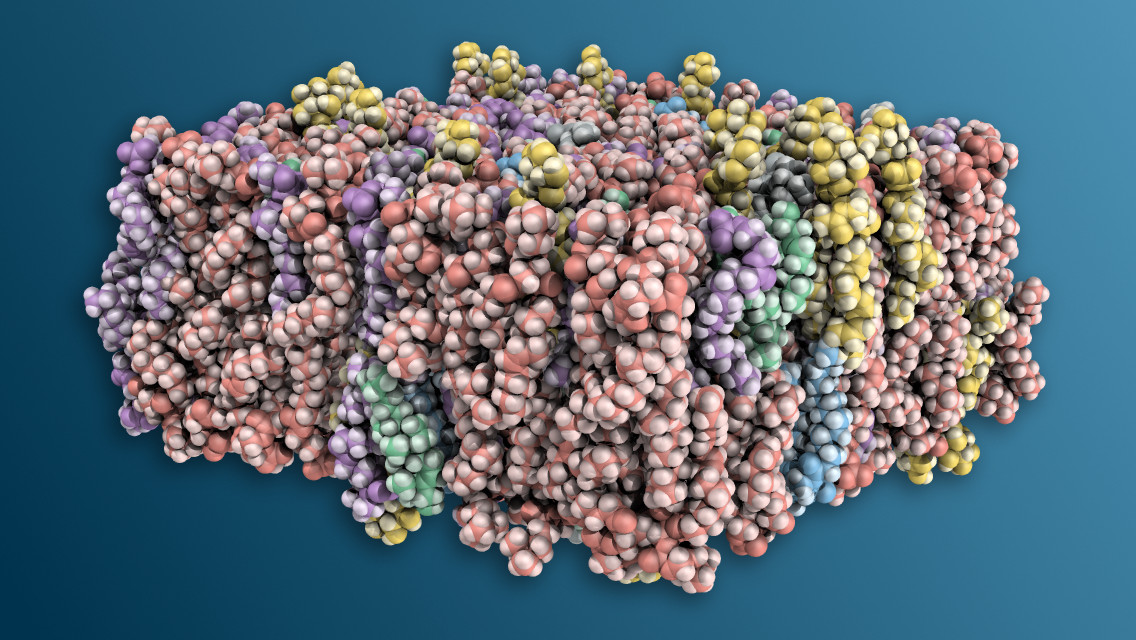
I teach the biochemistry section of an undergraduate general biology course and one of the topics I cover is cellular membranes. I always show the students the key molecular components and their interactions when covering this topic, and I have not found suitable molecular illustrations of membranes for all the aspects of membrane structure that I want to cover. I found an online membrane builder that will create files suitable for import into the popular open source molecular graphics software program, Pymol. Here is how I made the membrane illustration that accompanies this article.
Gather Membrane Lipid Composition Information
I wanted to model an endoplasmic reticulum (ER) membrane, so I roughly estimated the ER membrane lipid composition using Figure 2 from van Meer et al. (2008) Nat Rev Mol Cell Biol. 9(2): 112–124. doi: 10.1038/nrm2330.
Build the Membrane
Go to the CHARM-GUI Membrane Builder, and follow the onscreen instructions.
Choose the Membrane Only System then click Next Step.
- Choose
Heterogeneous Lipid - Box Type:
Rectangular - Length of Z based on:
Hydration numberand use37waters per lipid molecule - Length of XY based on:
Numbers of lipid components - XY Dimension Ratio:
1(the required value) - Sterols1:
- cholesterol
- # of Lipid on Upperleaflet:
19 - # of Lipid on Lowerleaflet:
19
- # of Lipid on Upperleaflet:
- ERG: leave the values at
0
- cholesterol
- PC (phosphatidylcholine) Lipids: 54%, 256 x 0.54 = 138.24, 138/2 = 69, choose
69POPC for each leaflet. - PE (phosphatidylethanolamine) Lipids: 30%, 256 x 0.3 = 76.8, 76/2 = 38, choose
38POPE for each leaflet. - PS (phosphatidylserine) Lipids: 2%, 256 x 0.02 = 5.12, 6/2 = 3, choose
3POPS for each leaflet. - PI (phosphatidylinositol) Lipids: 12%, 256 x 0.12 = 30.7, 30/2 = 15, choose
15SAPI for each leaflet. - SM (sphingo) Lipids: 2%, 256 x 0.02 = 5.12, 6/2 = 3, choose
3PSM for each leaflet.
Go back to the top of the page and click Show system info.
Go to the bottom of the page and click Next Step.
On the next page, leave the default values, and click Next Step.
When the page is done loading, click Next Step.
Continue clicking Next Step until a file named step5_assembly.pdb is generated. Download that file and rename it ER-membrane.pdb.
Color the Membrane Lipids in Pymol
I use Pymol to manipulate .pdb files. I made a script that will color each lipid species a different color. I have attached the script as a GitHub Gist, below.
Move the ER-membrane.pdb file and the pymol-ER-membrane.txt file into your Pymol working directory. Run the script from the Pymol command line using @pymol-ER-membrane.txt. The script will write a ray traced .png file to your working directory. Change the script as you see fit.
Note that the lighting settings to get a nice ray traced image produces a washed out image in the pymol viewer.
Finishing Touches
First, I uncommented the line set ray_opaque_background, 0 and ran the script again. This produces an image that has a transparent background. Then I imported the image into Affinity Designer and added a drop shadow and changed the background gradient to diagonal. Next, I exported it as a .jpg file.
Final Thoughts
Now that I have a membrane .pdb file, I can illustrate almost anything I want about membrane structure. I can color the membranes by hydrophobicity, label all the polar head groups, or show only specific lipids. I can zoom in and show packing of saturated vs unsaturated fatty acid tails, and I can show how sterols fit into the membrane. Pymol makes it easy to generate movies, if I want to go that far. Finally, I can always return to CHARM-GUI and build another membrane if I want to show membranes of other lipid composition.
1 The cholesterol to phospholipid ratio for the ER membrane is 0.15. For a membrane with 256 phospholipid molecules, one needs 256 x 0.15 = 38.4 cholesterol molecules, or 19 for the upper and lower leaflets. I know that each leaflet can have a different lipid composition, but since I don’t have that information handy, I’ll make the membrane symmetrical. I chose 256 for the number of phospholipids because it gives a membrane patch of a nice size. ↩